Previous: Complex Systems
Vacuum simulations are not realistic as proteins live in aqueous environments (and as time goes on, you will learn that “aqueous” is at best an approximation!). In order to simulate proteins more realistically, we must add water. There are many water models available; one of the most venerable is the TIP3 model. We will add TIP3 waters to our protein by using a plugin. In addition, solvation “by hand” using tkcon will be demonstrated.
Using the solvate plugin GUI
The solvate extension will automagically take your PSF/PDB pair, add TIP3 waters to a specified box size, generate all the segment information (new PSF and PDB), and load the new files into your VMD. When it works, it works quickly and easily. When it doesn’t, there are additional things we can try...
Make sure the only molecule you have loaded is your simulation PSF/PDB pair (and that these files gave you a successful simulation), and open the solvate plugin (Extensions..Modeling..Add Solvation box). The first thing you should notice is your PSF and PDB are listed in the first two lines of the window:
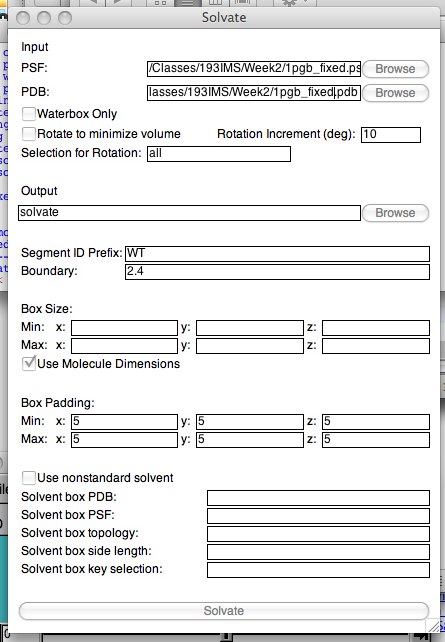
“Waterbox only” will leave your protein out of the generated PSF/PDB (so don’t click that!). You have the choice to rotate your molecule (in order to save space in the water box), and name your water segment (but you are restricted to four characters --- “WAT” is OK but “WATER” is not!). Make sure “Use Molecule Dimensions” is checked under the “Box Size”, and add some padding (at least 5A, but remember more padding = more water = more calculation time). The bottom section is not needed. At this point, when you click on the bottom “Solvate” button, a new PSF/PDB will be created and loaded into VMD.
If an error results, it is probably because your protein is not centered in the box, and ends up hanging outside the water box that solvate is trying to generate. In this case, follow this link to the orient plugin and run the script in order to center your molecule, write out a new PDB (“writepdb file.pdb”), then repeat the solvate steps above.
We have found the Solvate plugin to be rather buggy! If this does not work for you, don't worry! Use tkcon (see below)!
Using solvate in tkcon
The solvate GUI is simply a quick way to issue the solvate commands. Thus you can also accomplish this in the tkcon. It is relatively simple (the PSF/PDB do not even need to be loaded):
package require solvate
solvate protein_sim.psf protein_sim.pdb -t 10 -o protein_solvated
That’s it! “-t” stands for the padding (in each dimension), and the “-o” specifies the output PSF/PDB filenames.
Running your simulation/Analysis
At this point, you can take your new solvated PSF/PDB and generate a NAMD config file. Be careful: you will still have to be sure the parameters specified in the file point at the match to the top_all27_prot_na.rtf - par_all27_prot_na.prm.